Analysis sent out every Monday
Axial: https://linktr.ee/axialxyz
Axial partners with great founders and inventors. We invest in early-stage life sciences companies such as Appia Bio, Seranova Bio, Delix Therapeutics, Simcha Therapeutics, among others often when they are no more than an idea. We are fanatical about helping the rare inventor who is compelled to build their own enduring business. If you or someone you know has a great idea or company in life sciences, Axial would be excited to get to know you and possibly invest in your vision and company . We are excited to be in business with you - email us at info@axialvc.com
Phenotypic screening
Drug discovery as we know today really started off with natural products and finding chemical matter that leads to a biochemical change or a change in physical appearance. Phenotypic screening focuses on identifying phenotypes (and rescues) of interest. The power of genomics and structure-based drug design has revolutionized many parts of drug discovery by flipping this process to start at a particular target or pathway; however, phenotypic screening is still useful in some contexts where the underlying biology hasn’t been fully characterized. As a result, it is an old technology still useful for new problems. Moreover, this approach is not only useful in drug development. Phenotypic screening could bring transformative products in consumer products among other markets - Revela is leading the way here.
In 2015, a team at Pfizer published a paper defining the key rules for a phenotypic screen:
Selecting physiologically-relevant cell types and models. Examples are iPSCs, organoids, and even animal models. In vitro models offer higher throughput versus in vivo models that might have more accuracy. Parallel Bio is the leader here.
Designing assays the are relevant to a disease
Defining assay endpoints that are similar to clinical endpoints. This is focused on using biomarkers or disease signatures that are matched to clinical samples.
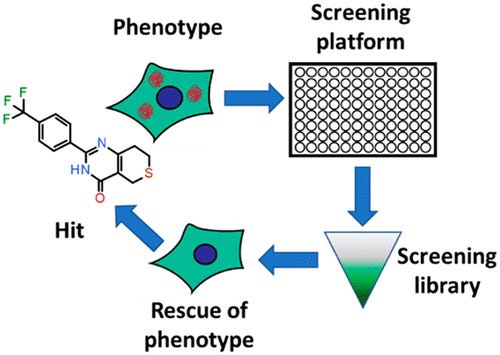
These types of screens can be divided into 2 major steps: (1) simple primary assay to cover as much chemical matter as possible and (2) complex, physiologically-relevant models to hone in on interesting hits. After the model is established, a large library of molecules are screened to measure something like expression changes in a panel of proteins or cellular characteristics like proliferation. Secondary assays are used mainly to counter screen and filter out molecules that have general effects. This is the part where phenotypic screening has higher upfront costs versus target-based programs. Automation of high throughput screening helps here as well as filtering hits more accurately with transcriptomics, proteomics, unsupervised machine learning, high-content imaging, and other tools within the assays themselves. Hits are then grouped into mechanism classes to prioritize them with a focus on target deconvolution. Even though many approved drugs have been approved without a known target, this knowledge is incredibly important to de-risk a program. Excitedly, this information is much more accessible with the wide-array of tools: panels of known target classes are screened against, activity-based protein profiling is very useful along with compound-immobilized beads, photoaffinity labeling, and cellular thermal shift assays.
In drug development programs, the key focus is formulating and testing a hypothesis. In target-based discovery, the process tests a hypothesis (biological, clinical, commercial) by generating a hit for a given target then going to lead selection and optimization and finally into pre-clinical/clinical testing. Whereas, phenotypic screening generates a hit for a given phenotype then the trick is to find the target. The latter process can get tricky sometimes and can make lead selection a bit more difficult, and as a result, better frameworks are needed to make phenotypic screening as structured as target-based programs:
In vitro models that are relevant for more diseases. Patient-derived cell lines are a step forward here, but could lead to unexpected variance in assay conditions. Co-culturing is also another useful approach but could impact throughput.
With more complex assay designs, identifying important variables that influence reproducibility and disease relevance.
“Chain of translatability,” an idea from biopharma, to connect assay endpoints to a clinical outcome.
What other tools are needed to advance phenotypic screening?:
Increasing the screening throughput of more complex models like organ-on-a-chip and organoids.
Translational work to build models for diseases that have recent breakthroughs in their mechanisms-of-action (MoA) and hopefully genetic drivers especially in CNS. On a side note, phenotypic screening is likely going to make a very large impact on longevity drug development.
More accessible proteomic tools particularly for activity-based profiling during the target deconvolution step. This would speed up the process and lower the barrier to entry to use phenotypic screening because this step is the last and sometimes hardest.
Simply, more case studies and work to merge phenotypic and target-based screening. The idea would be to create a database enabling a phenotypic match from a virtual screen one day.